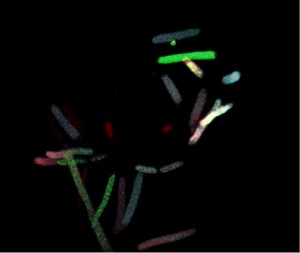
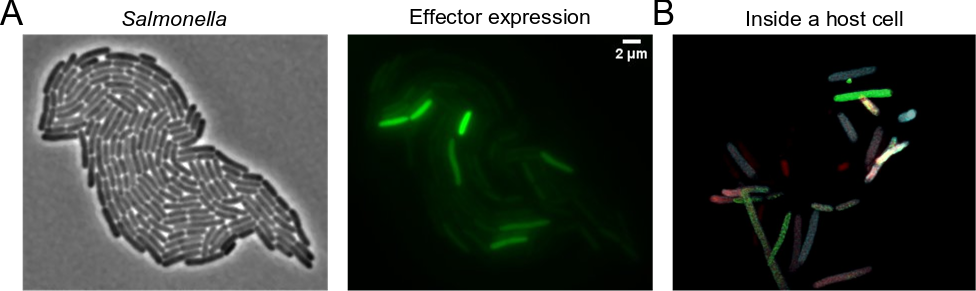
Salmonella
About 30 proteins are needed to create a fully functional T3SS machinery, which can deliver many different effector proteins directly into the cytosol of host cells. Together, genes necessary for proper function of the secretion system and its effectors constitute several per cent of total bacterial genome and their expression constitutes considerable metabolic burden. Although production of the T3SS and translocation of its effectors is beneficial in general, not all bacteria residing in the same conditions in the host express the secretion system and all its effectors thus forming a highly heterogeneous population of isogenic bacteria. This allows to split the metabolic burden between individual bacterial cells and to benefit from the metabolic cost paid by the other bacteria. We use the model organism Salmonella enterica to study the molecular mechanisms regulating the heterogeneity in expression of the T3SS and its effectors, and the physiological implications of this heterogeneity.